Molecular Nano-Engineering Lab.
TANII Research Group
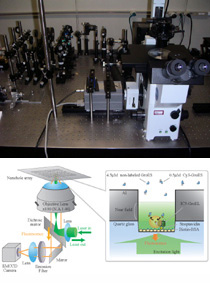


Single-Molecule Fluorescence Imaging Using Nanostructure Array
Real-time imaging of single molecule fluorescence using nanostructure arrays was achieved through collaborative efforts involving Waseda University and the University of Tokyo (Prof. Takashi Funatsu's group). The new method makes it possible to visualize binding release events of individual molecules at intracellular concentrations in real time. To date, we have succeeded in visualizing individual co-chaperonin GroES binding with chaperonin GroEL at 5 micro molar in real time (see the movie in this page).
The direct observation of binding release events at a high concentration indicates the possibility of analyzing weak interactions because molecules with a low association constant only bind frequently with the protein at a high concentration. While such weak interactions sometimes play important roles in living cells, the conventional method with diffraction-limited optics and total internal reflection light cannot identify the fluorescence signal from the molecule of interest at high concentrations due to strong background fluorescence from the large quantity of molecules within the observation volume.
The conventional method limits concentrations to 50 nano molar. To overcome this limitation, we investigated a new method that takes advantage of the nanostructure array fabricated on the glass slide. The nanostructure array, consisting of subwavelength waveguides in a thin metal film (e.g. zero-mode waveguide, near-field probe array, etc.), confines the excitation light to zept litter level, and the reduced excitation volume makes it possible to observe individual molecules at a high concentration with high signal-to-background ratio.
Demonstrations using GroES and GroEL have shown that real-time imaging of single molecule fluorescence is feasible even at micro molar levels, and the evaluated dissociation rate constants agree well with results reported to date, indicating that our system can be used to analyze protein functions revealed only at intracellular concentrations and offers the potential to resolve functions of various proteins.
Currently, we are trying to observe dynamics of cell membrane proteins and interaction between ligands and aptamers by modifying the nanostructure geometry. Numerical simulation suggests that the signal-to-noise ratio of real-time single-molecule fluorescence imaging can be enhanced by modifying the nanostructure geometry. This indicates that the real-time single molecule fluorescence imaging can be performed at a much higher concentration, visualizing interaction and dynamics of wide variety of proteins.
T.Miyake, T.Tanii, H.Sonobe, R.Akahori, N.Shimamoto, T.Ueno, T.Funatsu, I.Ohdomari: Real-Time Imaging of Single-Molecule Fluorescence with a Zero-Mode Waveguide for the Analysis of Protein-Protein Interaction, Anal. Chem. 80, 2008, 6018.
T.Sameshima, R.Iizuka, T.Ueno, J.Wada, M.Aoki, N.Shimamoto, I.Ohdomari, T.Tanii, T.Funatsu: Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zero-mode waveguides, J. Biol. Chem. 285 (2010) 23159.
T.Tanii, R.Akahori, S.Higano, K.Okubo, H.Yamamoto, T.Ueno, T.Funatsu: Improving zero-mode waveguide structure for enhancing signal-to-noise ratio of real-time single-molecule fluorescence imaging: A computational study, Phys. Rev. E 88, 012727 (2013)